XDS AUTOPROCESSING
The automated scripts run the programs XDS (data reduction, including integration and scaling), pointless (further space group determination), and aimless (final merging of data to produce the mtz you will need for structure solution). You can see the results directly within the b4 gui.
Pointless runs with default input and aimless uses the ONLYMERGE input. Aimless runs three times. For the first run, it produces the file aimless_all.log and aimless_all.mtz . As part of this run, it identifies the resolution cutoff based on cc(1/2)>0.3 and IsigI>2.0. It then runs again based on a resolution cutoff of cc(1/2)>0.3 and produces the files aimless_cc.log and aimless_cc.mtz . It then runs a final time based on IsigI>2.0, and produces the files aimless_IsigI.log and aimless_IsigI.mtz . Look for these files in the XDS folder which appears in your data directories. (Note that pointless and aimless only run when XDS has successfully produced the scaled file XDS_ASCII.HKL)
If processing went smoothly, you should see these files in your directory. You can use any of the mtz files for further structure solution:
- aimless_all.log This is the log resulting from autoprocessing using all the data.
- aimless_all.mtz This is the mtz file resulting from autoprocessing using all the data.
- aimless_cc.log This is the log file resulting from autoprocessing using a resolution cutoff based on a cc1/2 of 0.3.
- aimless_cc.mtz This is the mtz file resulting from autoprocessing using a resolution cutoff based on a cc1/2 of 0.3.
- aimless_IsigI.log This is the log file resulting from autoprocessing using a resolution cutoff based on IsigI > 2.0.
- aimless_IsigI.mtz This is mtz file resulting from autoprocessing using a resolution cutoff based on IsigI > 2.0.
- XDS.INP This is the input file used for processing in XDS. Here is a LINK which shows the parameters used in this file and what they all mean.
- XDS_ASCII.HKL This is the scaled reflection file from XDS. Note here that if XDS does not finish because of an issue with the data, you will not see this file in the directory. One thing to try in this case is to edit XDS.INP and change the line which reads JOB=XYCORR INIT COLSPOT IDXREF DEFPIX INTEGRATE CORRECT to: JOB=DEFPIX INTEGRATE CORRECT and then rerun XDS. (We have seen XDS fail when those first four entries in that line are present, but successfully process when they are not.)OTHER XDS SPECIFIC FILES OF INTEREST:
- COLSPOT.LP Gives the specifications used (and the results) for picking spots for indexing.
- CORRECT.LP Gives the specifications (and results) for scaling the data, including R-factor, number of observations, unique reflections. Gives a table towards the end of the file which is similar to the one output by HKL2000.
- DEFPIX.LP. Defines the “trusted” region of the detector.
- IDXREF.LP Gives results from indexing. Similar to the table given by HKL2000 when indexing.
- INIT.LP Shows the detector gain and dark current corrections used.
- INTEGRATE.LP Output file from integration.
- INTEGRATE.HKL The reflection file output.
- GXPARM.XDS The final refined cell parameters and orientation.
- SPOT.XDS Gives list of reflections.
- XPARM.XDS The orientation matrix file.
- XPLAN.LP Data collection strategy information.
DIALS AUTOPROCESSING
We are now running an automatic data processing using xia2 to run DIALS. Similar to xds, it creates a DIALS subdirectory in the data collection directory where the images are. Look at xia2.html file for a nice table showing the processing results. If you open the xia2.html in a browser, you will see the results in a table like this:
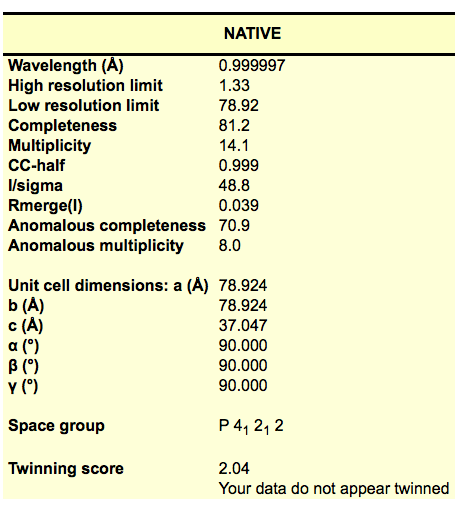
Note that DIALS often takes up to half an hour after the last image was collected to finish, so you won’t see the directory appear right away.
Within the DIALS directory, you will see another directory called DataFiles. This contains the sca and mtz files resulting from processing.
ARCHIVED:
Previously, we had automated scripts which processed data on 8.2.1 and 8.2.2 using Xia2, and the results were placed in the user home directory under the directory ~/rabbit_proc. We are no longer supporting these scripts, but you may still see some old processed data under this directory.